Project details
Acronym: MUSECoV
Researcher: Prof Dr Sophie le Poder
Countries: France, Italy, Spain, Poland
Website: www6.jouy.inrae.fr

Project Summary
Coronaviruses (CoVs), which are currently subdivided in four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus and Deltacoronavirus, infect numerous mammalian and avian species. Human CoVs and mammalian CoVs cluster into the Alphacoronavirus and Betacoronavirus genera. Avian CoVs belong to Gammacoronavirus and Deltacoronavirus genera. Some mammalian CoVs, infecting pigs in particular, are also described in the Deltacoronavirus genus. CoVs seem to have a significant resistance outside the host despite their nature as enveloped viruses, they possess the second largest known viral RNA genome, they are the only RNA viruses known to have an exoribonuclease function, and their replication mechanisms give them the potential for recombination.
The current pandemic “COVID-19” is the most important since the H1N1 Flu human pandemic of 1918. The causal viral agent of COVID-19, a human coronavirus, named SARS-CoV-2, probably originated from horseshoe bats which first crossed the species barrier into, an as yet, unidentified intermediate animal host.
A number of emerging diseases have resulted from coronaviruses crossing the species barrier such as SARS-CoV, MERS-CoV, and in veterinary medicine the SeACoV of pigs, (originating from bat HKU2-CoV).
Due to the potential of CoVs to cross the species barrier the first aim of the project will be to evaluate the circulation of SARS-CoV-2 in companion and domestic animals and to perform in-vitro studies that analyse its potential to replicate in cells derived from different animals. We will also address the cross-species potential of other coronaviruses from wildlife and domestic animals. To investigate these questions, we propose a consortium involving teams from France, Spain, Italy and Poland. It will ensure the collection of thousands of samples from diverse animals and different geographic locations. Using standardized RT-qPCR and serological analyses, the eventual diffusion of SARS-CoV-2 in domestic animals in different European countries of the consortium will be evaluated. The functional in vitro assays will complete these investigations and focus on the mechanistic aspects such as receptor function and specific intracellular host determinants.
The second aim of the project is to better understand the global circulation of animal CoVs and their genetic evolution dynamics under different constraints and ecological context. Samples collected from bats, domestic carnivorous, ruminants, poultry and wildlife over time in different European locations, will allow the analysis of their evolution rates at a multi-scale level (temporal and geographic), under natural conditions. These studies will be complemented by in vitro assays that will evaluate the frequency of recombination between different virus strains of the same species (avian or pig) when inoculated simultaneously onto cell cultures (Figure 1).
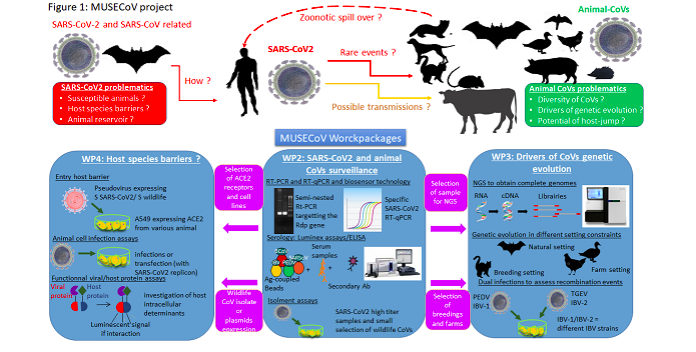
The multidisciplinary ambition of this project has brought together researchers involved in a wide range of disciplines: human virology (Partner 2), veterinary / animal virology (Partners 1, 2, 3 and 5), evolutionary biology (Partner 2), ecology (Partner 1, 2 and 5), molecular epidemiology (Partners 1, 2, 3 and 5), innovative methods and nanotechnology (Partner 2 and 4) and biochemical competences (Partner 3).
Such a team of specialists guarantees the acquisition of samples relevant to the interpretation of epidemiological and phylogenetic analyses. The current urgency as well as the innovative quality of the MUSECoV project will prove attractive to European experts on different coronaviruses, and enable complex cross-referencing and data verification within the consortium established.
The data generated by MUSECoV will be invaluable for a better comprehension of how coronaviruses evolve and circulate in European domestic and wild animals. These findings will help improve countermeasures and better prevent future coronavirus emergence in animals and humans.
